Rythms in time, patterns in space. How do living systems do it. What equations describe these phenomena? From the vein pattern in higher plants, through the circannual rythms of seasonal bulb floweing right upto schooling patterns of fish.
Friday, December 19, 2008
Neuronal model contest
Tuesday, September 30, 2008
Alignment of Polarized Cu Sticks
Some recent exciting research from the IISC Bangalore has experimentally demonstrated the breakdown of the central limit theorem that the sample-sample fluctuation of N particle systems grows as N^1/2. They demonstrate that for non-equilibrium systems the fluctuation can vary as N for N particles.
Interestingly their ordering of the Copper rods only occured in the driven system (between two vibrating plates) when the rods were etched at one end, effectively making them arrow-like. Here are some of the comments from Science:
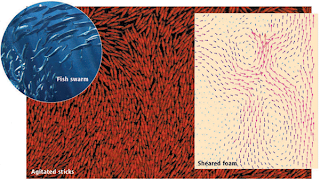
(FIGURE: Swarms and swirls. In the experiments of Narayan et al., agitated sticks form swarming states that exhibit giant number fluctuations. Similar patterns are observed in fish swarms (top left). Swirls are also observed in systems that are close to jamming, for example, in the motion of bubbles in a sheared foam (right).)
Science 6 July 2007: |
MATERIALS SCIENCE:
Shape Matters
Martin van Hecke
"Swarming and giant number fluctuations are a hallmark of the alignment displayed by driven collections of nonspherical particles. Theoretical models have been developed to describe swarming and alignment observed in schools of fish, flocks of birds, herds of sheep, or bacterial colonies--often borrowing from equilibrium models for magnetization, which consider the alignment of arrowlike objects. A very simple nonequilibrium model that exhibits cooperative motion arises when these arrows are allowed to propagate (2). In similar models, a collective response to predators and decision-making can arise (3). Toner and Tu first pointed out the giant fluctuations in such models (4).
In these systems, the particles have a preferred direction of propagation--just like real fish and birds. In 2003, Ramaswamy et al. (5) wondered what would happen for "active nematics," liquid crystals in which the particles have an orientation but have identical heads and tails (like the sticks in the present experiment). Their theory predicted that nematic systems also should exhibit giant number fluctuations, and these were recently observed in computer simulations (6).
However, when Narayan et al. tried to find such fluctuations in experiments, they encountered a surprising hurdle: cylindrical rods, arguably the simplest nematic particles, do not form nematic states and do not exhibit giant fluctuations (7). The authors achieved their present breakthrough only after etching the rods to obtain sticks with thinner ends (see the figure); for unknown reasons, these sticks exhibit nematic order. To complicate matters further, Aranson et al. recently performed similar experiments and observed that weak coupling between the nematic order and spurious in-plane vibrations of the support plate may strongly influence the swirling motion (8). Clearly, swarming is a subtle problem, and the precise nature of the swarming state and the transition to swarming is not yet fully understood.
The experiments of Narayan et al. are part of a bigger story, where nonequilibrium systems of nonspherical particles exhibit surprising behavior: We do not yet understand the consequence of shape. An earlier striking example of this is the finding that, contrary to expectation, M&M candies can be packed more effectively than spheres (9)."
REFERENCES:
- V. Narayan, S. Ramaswamy, N. Menon, Science 317, 105 (2007).
- T. Vicsek, A. Czirok, E. Ben-Jacob, I. Cohen, O. Shochet, Phys. Rev. Lett. 75, 1226 (1995).
- I. D. Couzin, J. Krause, N. R. Franks, S. A. Levin, Nature 433, 513 (2005).
- J. Toner, Y. Tu, Phys. Rev. E 58, 4828 (1998).
- S. Ramaswamy, R. A. Simha, J. Toner, Europhys. Lett. 62, 196 (2003).
- H. Chaté, F. Ginelli, R. Montagne, Phys. Rev. Lett. 96, 180602 (2006).
- V. Narayan, N. Menon, S. Ramaswamy, J. Stat. Mech. 2006, P01005 (2006).
- I. S. Aranson, D. Volfson, L. S. Tsimring, Phys. Rev. E 75, 051301 (2007).
- A. Donev et al., Science 303, 990 (2004).
- A. J. Liu, S. R. Nagel, Nature 396, 21 (1998).
- O. Dauchot, G. Marty, G. Biroli, Phys. Rev. Lett. 95, 265701 (2005).
- A. S. Keys, A. R. Abate, S. C. Glotzer, D. J. Durian, Nature Phys. 3, 260 (2007).
- W. G. Ellenbroek, E. Somfai, M. van Hecke, W. van Saarloos, Phys. Rev. Lett. 97, 258001 (2006).
- F. Lechenault, O. Dauchot, G. Biroli, J.-P. Bouchaud; available online at http://arXiv.org/abs/0706.1531v1.
Wednesday, August 06, 2008
Spring cleaning the WinDisk (win doof auf deutsch)
Here are some tips from the trawling of the net.
- Delete everything inside the Windows Prefetch folder (\\Windows\Prefetch) - you may save a couple of MBs and may also notice that Windows loads faster. (ref1).
- Clean the cache as follows:
----------
Cleaning the cache
If you use Internet Explorer, cleaning out the cache is relatively easy. You can do it in a couple of different ways. If you have the browser open, follow these steps:
1. Choose Tools --> Internet Options.
IE displays the Internet Options dialog box.
2. On the General tab, click the Delete Files button.
A warning dialog box offers to delete all your offline content, as well. For most folks, this doesn't matter — they don't browse offline. If you're in the minority that does, select the check box.
3. Click OK.
IE deletes the cache files. Depending on how cluttered your system is, this can take a while.
4. When IE is done deleting cache files, click OK to close the Internet Options dialog box.
You now have a fresh, clean cache, ready once again to be cluttered with new pictures from your Internet road trips.
You can also clean out the cache without even opening the browser — just use the Disk Cleanup tool:
1. Choose Start --> All Programs --> Accessories --> System Tools --> Disk Cleanup.
The Disk Cleanup tool starts running. If you have multiple hard drives on your system, you're asked to choose which hard drive to analyze.
2. Pick a hard drive and click OK.
Disk Cleanup looks through your hard drive, calculating how much space it can reclaim. This process may take a while. A dialog box indicates items you can clean.
3. Make sure the Temporary Internet Files option is selected.
Temporary Internet files translates to what's stored in the cache. You can also pick other things to clean up, if desired.
4. Click OK.
Disk Cleanup displays a dialog box asking if you want to proceed.
Depending on what you asked Disk Cleanup to do, the actual cleanup can take a few minutes to complete. Cleaning out cache files this way takes no longer than cleaning from within the browser.
Finding the cache
Most people never worry about where their browser stores its cache files. Microsoft recognizes this and doesn't make a big deal of advertising where the cache is located. You can locate the cache by displaying the Internet Options dialog box (in the browser, click Tools --> Internet Options) and then clicking Settings. The resulting Settings dialog box indicates where the cache is located (next to Current Location in the middle of the dialog box).
As you examine the path name, notice that it's associated with the current user. If your computer is shared by multiple users, Windows creates a cache folder for each.
Another interesting tidbit is that Microsoft hides the cache folder. It doesn't hide it in the Settings dialog box, but it does hide it if you try to get to the folder yourself.
To see how this works, open a My Computer window for the C: drive. Double-click the Documents and Settings folder, then your account name. If you don't see a Local Settings folder in the account folder, even though the Settings dialog box says you should, don't worry. The reason is that the Local Settings folder is configured as a hidden folder; it doesn't show up when normally viewing folders.
To see the hidden folder, follow these steps:
1. In the folder window, choose Tools --> Folder Options.
Windows displays the Folder Options dialog box.
2. In the Advanced Settings area on the View tab, select the Show Hidden Files and Folders option.
3. While you're at it, deselect the Hide Extensions for Known File Types and also the Hide Protected Operating System Files options.
(You might have to scroll down the list to see these options.) Deselecting these two options ensures that you see as much information as possible; with more information, you can make better decisions in the long run.
4. Click OK.
As the Folder Options dialog box disappears, the information in the account folder is updated. You can now see the Local Settings folder, but the folder icon should appear a little more washed out than other icons. This indicates that the folder is normally hidden. You can still double-click it and then double-click the Temporary Internet Files folder to see what's in your cache.
Changing the cache size
Internet Explorer makes a point of ensuring that your cache never gets too big. In some respects, the cache is similar to the Recycle Bin — when its maximum size is attained, it starts deleting the oldest files to make room for the new files.
The problem is that the cache is often set much larger than it needs to be. The Settings dialog box has a control that indicates the amount of disk space to use for the cache. By default, Internet Explorer allocates 10 percent of your disk to its cache folder. This doesn't mean the folder automatically uses that much space, just that IE keeps storing files in the cache (without deleting any of them) until that 10 percent mark is reached.
Think about that for a moment — most computers these days come with at least a 20-, 40-, or 60GB hard drive. If all that space is allocated to a single drive and IE uses that drive for its temporary files, you can easily set a maximum cache size to 2-, 4-, or 6GB. Wow. If the average image downloaded from a Web site is approximately 10K, IE could store anywhere from 200,000 to 600,000 images on your system without deleting anything. Talk about clutter! When hard drives were smaller, the 10 percent rule made more sense.
You can lower the cache size by following these steps:
1. In the browser, choose Tools --> Internet Options.
The Internet Options dialog box appears.
2. On the General tab, click the Settings button.
The Settings dialog box appears.
3. Adjust the cache size via the Amount of Disk Space to Use slider or by typing a number in the text box.
You shouldn't hesitate to lower the cache space to 75MB. If you have a high-speed Internet connection, lower it even more — perhaps to 35MB or 40MB.
By making this simple adjustment, you save lots of hard drive space for better uses and won't hurt the overall performance of Internet Explorer.
(ref2)
----------
References:
1. Don't Prefetch
2. How dummies cleanup cache no catches here
Tuesday, June 24, 2008
ImageJ and Elcipse for plugin development
With the aim of segmenting some slightly complicated radial data, I decided the existing algorithms and tools were not sufficient. So instead of doing it the easy way- using MATLAB's ImageProcessing toolbox, I decided to go the open-source way. And its in Java and nobody who wants to try using my tool needs anything more than an internet connection and a machine capable of running imageJ.
Since I have found programming, particularly in Java using Eclipse to be such a pleasure I decided to go that way. A nice little introduction to how to configure and ImageJ plugin project in Eclipse (introduction to Eclipse elsewhere) was available, I have summarised the settings as briefly as possible:
1. When creating File->New->Java Project, click 'Make separate source and output folders'.
2. Rename the output folder 'plugins'
3. Put source files or make new ones in the src folder.
4. Create a new folder 'Externals' for external Jars that you would need (like JAMA- numerical package for matrices).
5. Copy ij.jar (the imageJ jar file) into the topmost layer of the project heirarchy.
6. Click on the project in the Eclipse window, then go to Project->Properties->Java Build Path->Libraries->Add External Jars and add those jars needed (ij.jar is a MUST, all others optional- depending on the plugin).
7. If a simple plugin is written in the src folder, run using the Run->Open Run Dialog
Here:
MAIN:
Project: myplugin
Main class: ij.ImageJ
ARGUMENTS:
Program arguments:
-Dplugins.dir=C:\Code\myplugin -Dmacros.dir=C:\Code\myplugin
VM arguments:
-Xms256m -Xmx512m
And then RUN!
Should work!
Tuesday, April 22, 2008
Getting to grips with Scientific Computing Tools on a MAC-osX

For somebody who spent time (and I am not proud of it) on a win32 machine and was quite happy even from a 64bit machine to keep programming 32bit, and sending off code to the big UNIX machines, its a bit of a mixed bag to be sitting at a Mac OS X terminal. MacBookPro. For one it needs a lot of beer.
Then it needs Octave - for which there is a binary installer available. But to plot anything you need gnuplot- anybody with Linux or Cygwin will tell you that- how on Mac I pray- well get the gnuplot precompiled file. Set the .profile files to set the path to the applications:
alias gnuplot="/Applications/Gnuplot.app/Contents/Resources/bin/gnuplot"
alias octave="/Applications/Gnuplot.app/Contents/Resources/bin/octave"
Oh, and as you might discover you have bash shell (mac OS X 10.5.*) but no .profile or .bash_profile file, then make it and add those lines to it. Kippis! Shanti.
Well. The Octave user list was VERY useful in fixing that. Here is the fix:
//------------------------
The alias command sets up a variable that applies only in your shell.
It won't help octave find gnuplot. You probably need to set up a symbolic
link to gnuplot in some directory on your path. Try:
ln -s /Applications/gnuplot/Gnuplot.app/Contents/Resources/bin/gnuplot \
/usr/local/bin/gnuplot
If this complains about permission precede this with a sudo and enter your
password when prompted.
//------------------------
"Mein f******, I can plot again", to rephrase Dr. Stragelove or Doktor Merkwuerdigliebe from Dr. Strangelove.
Wednesday, April 09, 2008
Compiling AgentCell
===========
# Note the file list.
filelist="c:/cygwin"$PWD"/filelist"
===========
All else remains the same. So the final javacompile file looks like this:
----------------------------------------------------------------------
#!/bin/bash -x
# Note the initial directory.
calldir=$PWD
# Note the file list.
filelist="c:/cygwin"$PWD"/filelist"
# Move to the initial directory.
cd $calldir
#DEFINE the win32 native java install using cygdrive reference to drive C:\
#JPATH=/cygdrive/c/Java/jdk1.6.0_05
# Find the available Java files.
find edu -name *.java -print > $filelist
find uchicago -name *.java -print >> $filelist
#show filelist
#cat $filelist
# Compile the available Java files.
javac -classpath .:lib/repast.jar:lib/colt.jar:lib/commons-collections.jar:lib/commons-logging.jar:lib/geotools.jar:lib/jakarta-poi.jar:lib/jgl3.1.0.jar:lib/jh.jar:lib/jmf.jar:lib/JTS.jar:lib/junit.jar:lib/log4j.jar:lib/mediaplayer.jar:lib/multiplayer.jar:lib/plot.jar:lib/sound.jar:lib/trove.jar:lib/xerces.jar:lib/hdf4.jar:lib/jhdf4obj.jar:lib/jhdf.jar:lib/jhdfobj.jar @$filelist
#javac -cp .:lib/repast.jar:lib/colt.jar:lib/commons-collections.jar:lib/commons-logging.jar:lib/geotools.jar:lib/jakarta-poi.jar:lib/jgl3.1.0.jar:lib/jh.jar:lib/jmf.jar:lib/JTS.jar:lib/junit.jar:lib/log4j.jar:lib/mediaplayer.jar:lib/multiplayer.jar:lib/plot.jar:lib/sound.jar:lib/trove.jar:lib/xerces.jar:lib/hdf4.jar:lib/jhdf4obj.jar:lib/jhdf.jar:lib/jhdfobj.jar:./edu/*:./uchicago/*
# Clean up.
rm $filelist
----------------------------------------------------------------------
Now to actually try to do the calculations the AgentCell bactertial paper did. Somehow my first tries aren't too promising! :) More later.
-------------------
However, it seems that the RePast libraries aren't being properly recognized. Since I had a good experience working with RePast and Eclipse as an IDE, decided to switch to that. Imported the source file structure. It compiles, and linked stochsim.dll with the following command to the VM:
-Djava.library.path=$libstochsimpath
Oddly the example doesn't compile asking for an output folder etc. So to all obvious purposes that part works.
Tuesday, April 01, 2008
Stochastic Reactions and Agent-Cells
I am now trying to compile the package (2008-04-01, 20:15:00) and see if I can modify it to my purposes. My goal is to levarage my experience (and joy) in working with agent based systems and model eukaryotic cellular processes with it. Possibly the stochastic simulator might be the right tool to address issues of stochasticity and multi-stability in reaction networks while at the same time studying spatial patterns!
The initial part is done. StochSim compiles with some flags changed in the makefile for CYGWIN under win32. Here are the modifications:
######################################################
# Options to build the release version with
# cygwin on windows assuming that C:\cygwin\BIN is on
# the path.
# If you are using eclipse, make sure to append
# C:\cygwin\BIN to the PATH in
# project->Properties->C/C++ Make->Environment
######################################################
# ### Shell
SHELL = /bin/sh
# ### Copy
CP = cp
# ### Remove
RM = rm -f
# ### Java
#JAVA_HOME = /cygdrive/c/Program\ Files/Java/j2sdk1.4.2_10
#JAVA_HOME = /cygdrive/c/Program\ Files/Java/jdk1.5.0_06
JAVA_HOME = /cygdrive/c/Java/jdk1.6.0_05
# ### Compiler
CXX = g++
OPTS = -D_STD_CPP -D_UNIX -D_DYNAMIC_UPDATE
CXXFLAGS = -mno-cygwin -O3 -Wall -Wno-deprecated \
-I$(JAVA_HOME)/include -I$(JAVA_HOME)/include/win32
OPTS = -D_DEBUG -D_STD_CPP -D_UNIX
CXXFLAGS = -g -mno-cygwin -Wall -Wno-deprecated \
-I$(JAVA_HOME)/include -I$(JAVA_HOME)/include/win32
# ### Linker
LD = g++
LDFLAGS = $(CXXFLAGS) -L$(JAVA_HOME)/lib -ljvm
#LDFLAGS = $(CXXFLAGS)
STOCHSIM_EXE = stochsim.exe
# ### Library
MKLIB = g++
LIBFLAGS = $(CXXFLAGS) -Wl,--add-stdcall-alias -shared
STOCHSIM_LIB = stochsim.dll
# ### Installation
BINDIR = ../bin/
LIBDIR = ../lib/
=============================================================
The agent-cell compile still doesn't work! But maybe the answer lies in the post from the AgentCell website for Win32 instructions:
Hi Iqbal,
AgentCell is written in Java so it will work on any platform. However it
includes StochSim that is written in C++ and therefore must be
compiled for the platform you are using.
I have never used AgentCell on windows. The way I would go about it is
as follows:
1) Install cygwin (http://www.cygwin.com/) on windows. Cygwin will
provide a "Linux" under windows. When you install cygwin make sure to
include the development tools. In particular the compilker gcc (or
g++)
(but not the java extensions to gcc: you want to use the native java
compiler from the Java SDK instead).
2) Open a cygwin prompt window and install the Linux version of Java
SDK
1.4 (http://java.sun.com/j2se/1.4.2/downloads/index.html) under Cygwin
3) Open a cygwin prompt window and install AgentCell following the
instructions we give for Linux.
Thierry
================= Maybe there be hope? ===============
We shall see.
I already have the latest java compiler from Sun, but its a Win32 install- therefore it understands only win32 native paths eg: C:\Java\etc. However Cygwin is Unix based and understands UNIX paths eg: C:/Java/etc. Further complicating the matter is a call from under Cygwin to the Windows javac.
The solution appears to be to use
cygpath -u for UNIX
cygpath -w for WIN
and to criss-cross as necessary. Lets see if this works.
The javacompile file is fairly mutilated by now!
References
- Athale C, Mansury Y, Deisboeck TS. (2005) Simulating the impact of a molecular 'decision-process' on cellular phenotype and multicellular patterns in brain tumors. J Theor Biol. 2005 Apr 21;233(4):469-81. Erratum in: J Theor Biol. 2006 Apr 21;239(4):516-7.
- Mansury Y, Kimura M, Lobo J, Deisboeck TS. (2002) Emerging patterns in tumor systems: simulating the dynamics of multicellular clusters with an agent-based spatial agglomeration model. J Theor Biol. 2002 Dec 7;219(3):343-70.
- Thierry Emonet 1,*, Charles M. Macal 2, Michael J. North 2, Charles E. Wickersham 1 and Philippe Cluzel (2005) "AgentCell: a digital single-cell assay for bacterial chemotaxis", Bioinformatics 21(11), 2714-2721 (online)